Using C. elegans to reveal how and why individuals differ in metabolism
2025-12-03 15:25
Keywords
Inter-individual metabolic variation, 3-hydroxypropionate (3HP), HPHD-1 genetic variation, Personalized metabolism model
Contribution of SunyBiotech
SunyBiotech is very honored to provide strain construction services for this study.
Strains:
PHX2627 hphd-1(syb2627) N2K172M,
PHX2628 hphd-1(syb2628) N2K172M,
PHX2640 hphd-1(syb2640) DL238M172K,
PHX2666 hphd-1(syb2666) DL238P268L,
PHX2667 hphd-1(syb2667) DL238P268L,
PHX2757 hphd-1(syb2757) DL238M172K
Introduction
Metabolic differences among individuals arise from a complex interplay of genetics, diet, microbiota, and environment. In humans, dissecting these factors is challenging because each person’s diet and lifestyle vary widely. Fox et al. (Nature, July 2022) addressed this problem by using the nematode Caenorhabditis elegansas (C. elegans) a controlled model to study metabolic individuality. C. elegans offers key advantages: it is a self-fertilizing hermaphrodite (allowing large, genetically identical populations), it can be fed a precisely defined diet, and it has a well-annotated genome with established metabolic network models.
Metabolic Profiling of Strains
The study compared four C. elegans strains with different genetic backgrounds (Figure 1a): the standard laboratory reference N2 strain (from Bristol, UK) and three wild isolates (BRC20067 from Taiwan, CB4856 and DL238 from Hawaii). N2 and BRC20067 are closely related, while CB4856 and DL238 are more divergent. All worms were synchronized at the first larval stage (L1) were grown in chemically defined K medium with lysed and lyophilized Escherichia coli HB101 as a diet to avoid the potentially confounding effects of active bacterial metabolism.
The researchers performed comprehensive metabolomic profiling on each strain’s endo-metabolome (cellular metabolites) and exo-metabolome (secreted metabolites). Many matched known metabolites (amino acids, fatty acids, nucleotides, etc.), but the majority were previously uncharacterized.
Comparisons between the wild isolates and N2 revealed several hundred metabolites with significantly different levels (Figure 1b-c). For example, an indole-containing glycoside dubbed iglu#93 (black arrows indicate) was abundant in all three wild strains but absent in N2 (Figure 1b). Several hydroxylated fatty acids were also enriched in the wild isolates (Figure 1d). The most dramatic difference among annotated compounds was in 3-hydroxypropionate (3HP): DL238 secreted over seven times as much 3HP into the medium as the average of the other strains (Figure 1e). 3HP is an intermediate in the propionate shunt pathway, which C. elegans uses when the normal vitamin B12-dependent breakdown of propionate is insufficient. The elevated 3HP in DL238 suggested a bottleneck in its propionate metabolism.
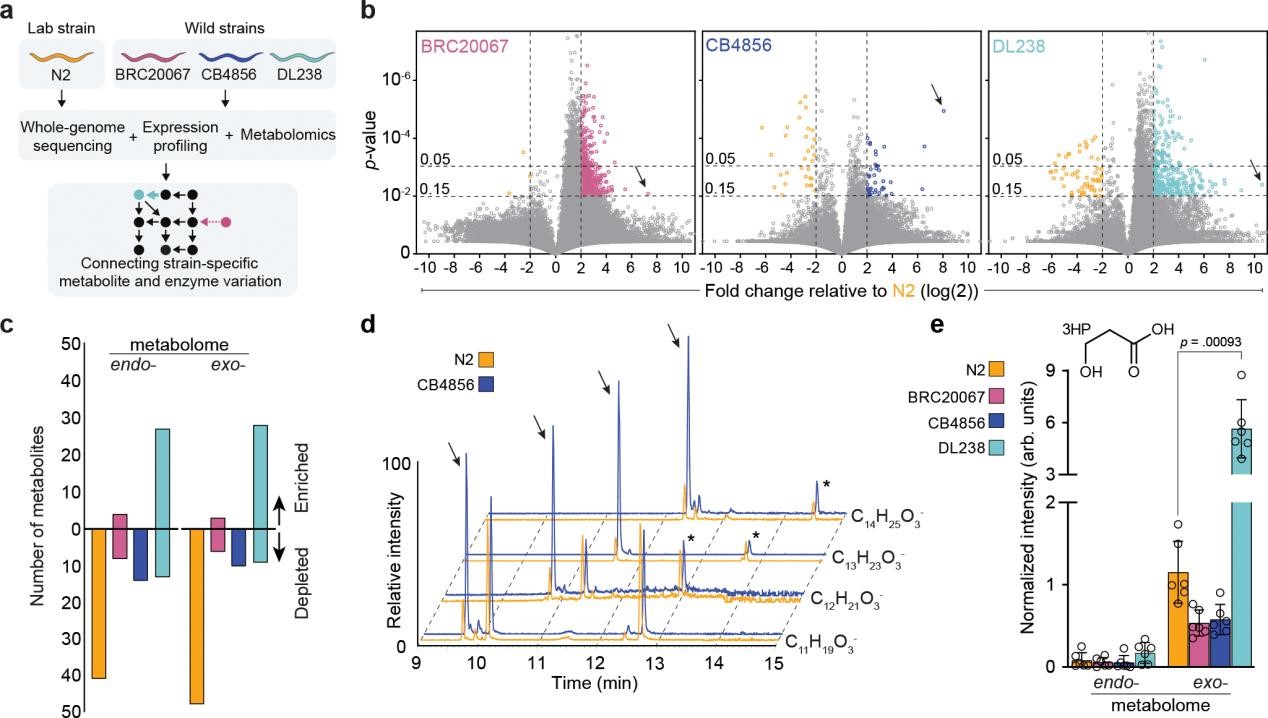
Figure 1. Inter-individual variation in metabolism
Discovery of 3HP–Amino Acid Conjugates
Many of the strain-specific features turned out to be novel metabolites. Tandem MS and molecular networking grouped these unknowns by structural similarity. This analysis uncovered a cluster of related compounds that were highly enriched in DL238’s exo-metabolome and scarce or absent in N2 (Figure 2a-b). Based on their exact masses and fragmentation patterns, Fox et al. identified these molecules as conjugates of 3HP (or a closely related hydroxy acid) with amino acids. In other words, they are 3HP–amino acid (3HP–AA) conjugates (for example, 3HP-Valine, 3HP-Leucine/Isoleucine, 3HP-Phenylalanine, etc.).
The appearance of 3HP–AA conjugates implied a shunt-within-a-shunt mechanism. The researchers reasoned that if the canonical (vitamin B12-dependent) propionate pathway were reactivated, the shunt intermediates would no longer accumulate, and the conjugates should disappear (Figure 2c). Indeed, supplementing DL238 with vitamin B12 (thus restoring the primary propionate catabolism) caused the 3HP–AA conjugates to drop dramatically (Figure 2d). 3HP is oxidized to malonic semialdehyde by HPHD-1, and hphd-1 disruption in the N2 background causes dramatically increased 3HP levels. Abundance of the putative 3HP-AA conjugates is also greatly increased in hphd-1(ok3580) mutant animals (Figure 2e). The researchers performed isotope tracing experiments with 13C5-Val in hphd-1(ok3580) mutants and the N2 strain to confirm the conjugation between 3HP and AAs (Figure 2f-g). This confirmed that these novel metabolites are produced only when 3HP accumulates because the usual breakdown route is impaired.
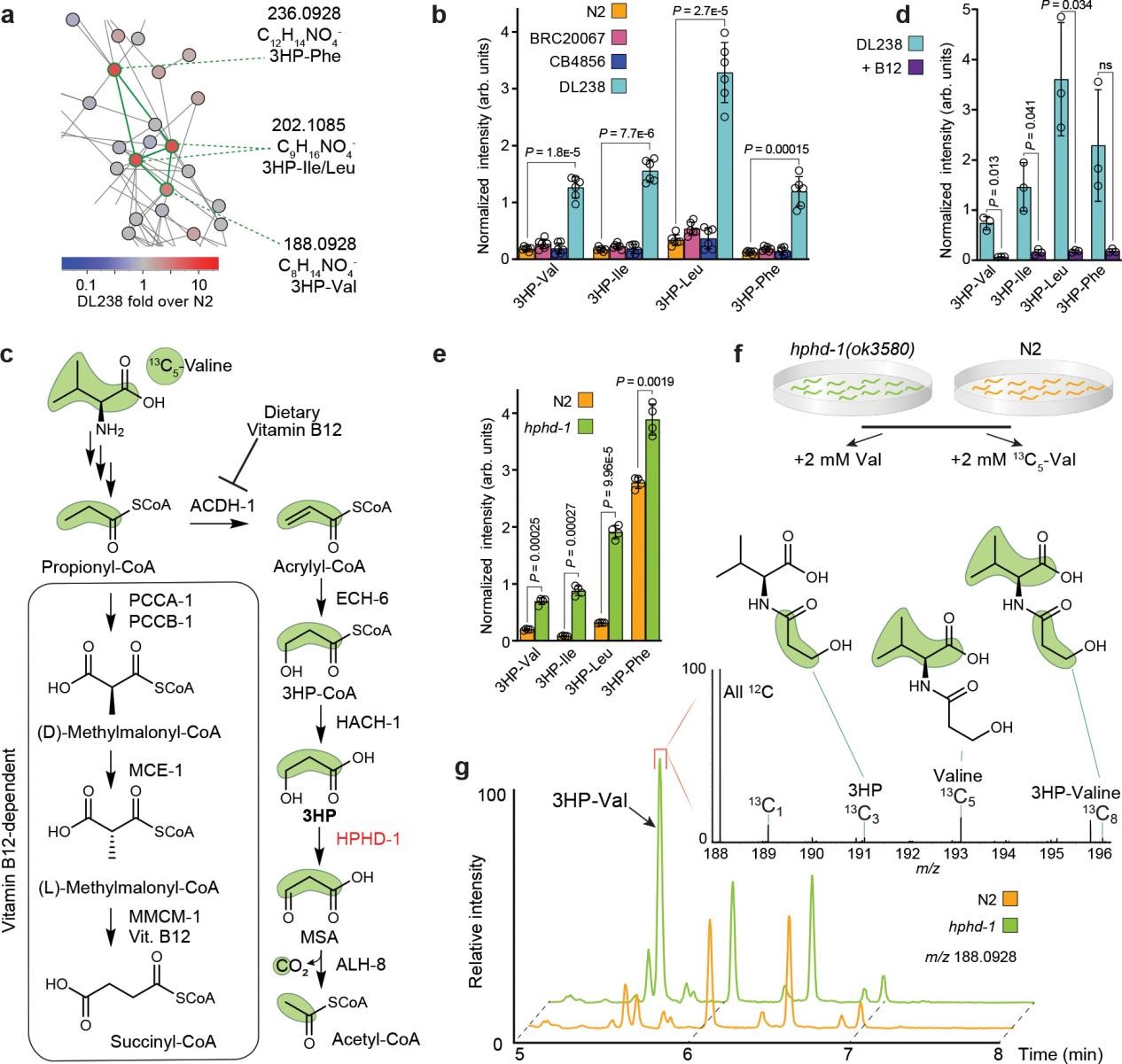
Figure 2. A shunt-within-a-shunt for propionate degradation
The Propionate Shunt and HPHD-1 Enzyme
Normally, propionyl-CoA is converted to succinyl-CoA via a B12-dependent pathway. Under B12 limitation, C. elegans uses a propionate shunt: propionyl-CoA is dehydrated to acrylyl-CoA, then reduced to 3-hydroxypropionyl-CoA, which is hydrolyzed to free 3HP. Finally, the enzyme HPHD-1 oxidizes 3HP to malonic semialdehyde, feeding into central metabolism.
DL238 carries two HPHD-1 variants (K172M and L268P) (Figure 3a). L268P is a reduction-of-function allele, which leads to the significant accumulation of 3HP and 3HP-AA. K172M has little effect in N2 but reduces 3HP/3HP-AAs in DL238, indicating context-dependent influence (Fuigure 3b-d). The finding that propionate is directed to N-propionyl-AAs in DL238 animals independent of the HPHD-1 edits indicates that propionate metabolism in DL238 exhibits additional differences compared to N2 (Figure 3e). Excess 3HP delays DL238 development more than N2, and this sensitivity maps to the single L268P substitution (Figure 3f-g).
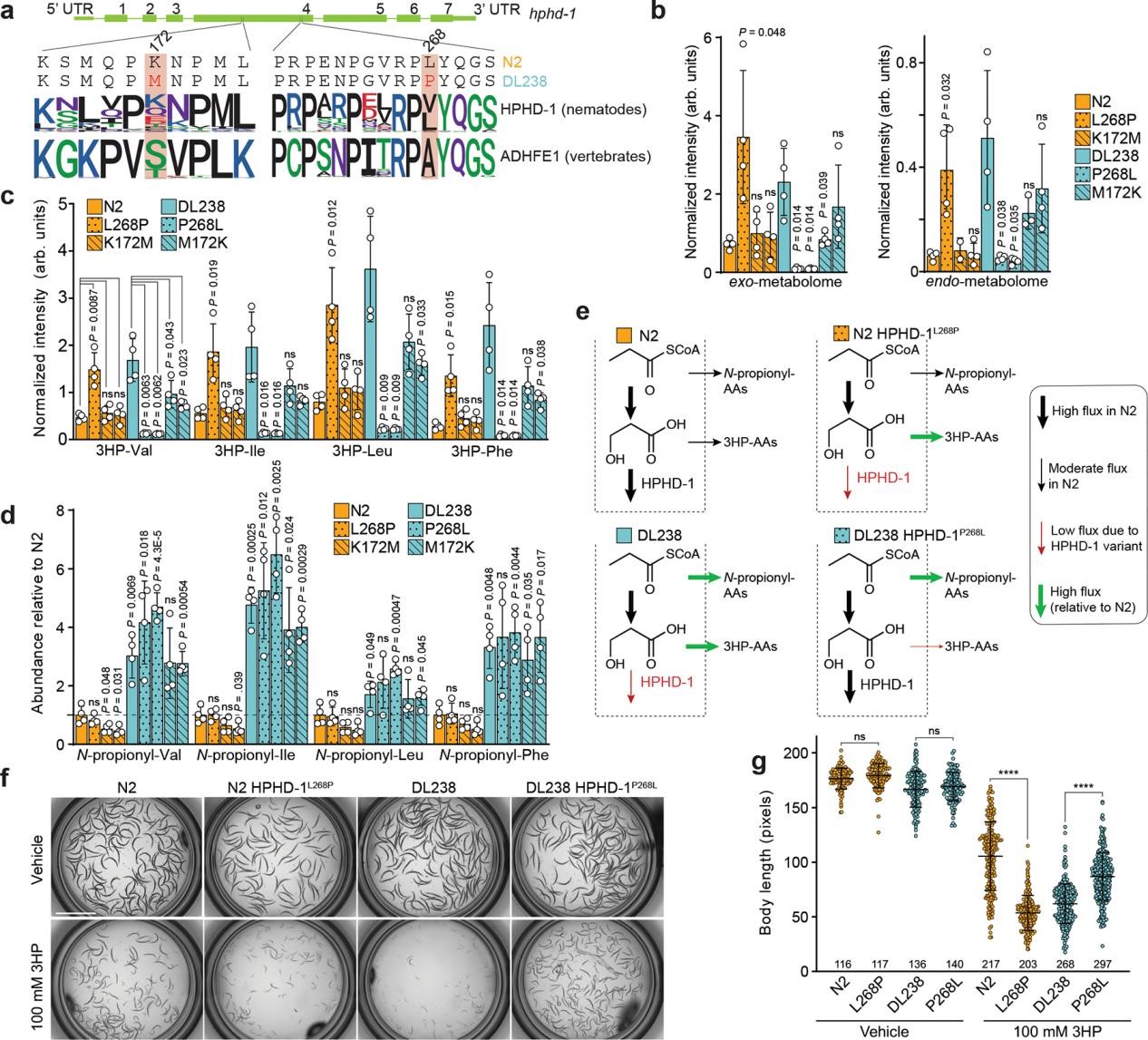
Figure 3. Genomic variation in hphd-1 causes 3HP and 3HP-AA accumulation in DL238
Gene Expression and Metabolic Network Modeling
Gene expression analysis showed that DL238 has higher transcripts for several propionate-shunt enzymes (hach-1, hphd-1, alh-8) compared to N2 (Figure 4a). This is consistent with a compensatory response: because HPHD-1 activity is weaker, the worm ramps up related enzymes to try to clear 3HP. This finding highlights an important point: in different genetic backgrounds, elevated expression of an enzyme can signal compensation for a defective allele, not necessarily increased metabolic capacity.
To integrate their findings into a systems-level framework, the authors expanded the C. elegans metabolic network model. They added new reactions to the existing iCEL1314 model to account for the formation of 3HP–amino acid and N-propionyl–amino acid conjugates, along with their transport (Figure 4b). The resulting pan-iCEL model begins to capture metabolic capabilities beyond the reference strain. As more strain-specific metabolites are discovered and characterized, this expanded model can grow, paving the way for metabolic network reconstructions that reflect species-wide genetic diversity.
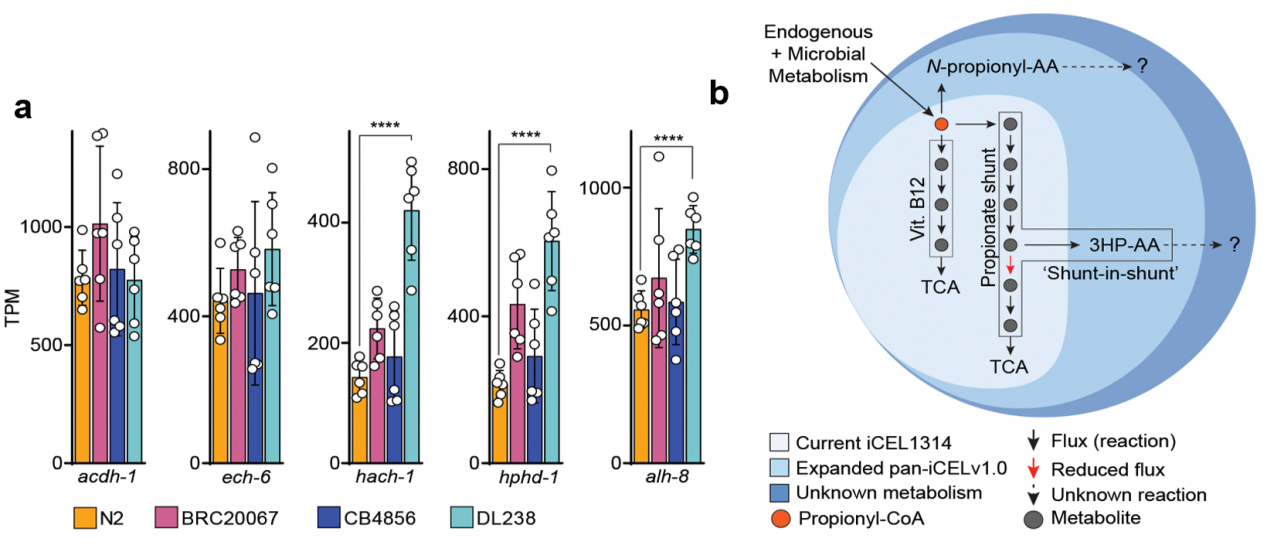
Figure 4. Propionate shunt gene expression and the iCEL1314 metabolic network model
Conclusion
The author's research has demonstrated that C. elegans can serve as a powerful model for linking natural genetic variation to metabolic phenotypes. By profiling multiple strains under tightly controlled conditions, the study uncovered not only expected metabolite differences but also entirely new compounds and pathways. The discovery of 3HP–amino acid conjugates and the identification of the HPHD-1 L268P variant driving their accumulation exemplify how comparative metabolomics and genomics can pinpoint the molecular basis of metabolic variation.
These findings have broader implications. Propionate metabolism is highly conserved, and its dysregulation causes human metabolic disorders (such as propionic acidemia). Dietary factors like vitamin B12 also influence propionate processing. The shunt-within-a-shunt adaptation uncovered here may represent a general strategy to detoxify accumulating intermediates when a primary pathway is compromised. More generally, this work lays the groundwork for personalized metabolic network models that incorporate genetic variation, with potential relevance to nutrition and disease in humans.
Reference
Fox BW, Ponomarova O, Lee YU, et al. C. elegans as a model for inter-individual variation in metabolism. Nature. 2022;607(7919):571-577.
doi:10.1038/s41586-022-04951-3






