FEM1C: A New Player in Neurodevelopmental Disorders
2024-08-23 10:21
Recent research has illuminated a novel genetic etiology for a severe neurodevelopmental disorder characterized by absent speech, pyramidal signs, and limb ataxia. The identification of a de novo missense variant in the FEM1C gene represents a significant breakthrough in our understanding of the complex genetic underpinnings of this condition. FEM1C, a less-studied component of the ribosome, has been largely overlooked in the context of human disease. This study by Dubey AA, et al. underscores the importance of exploring the functional roles of even the most seemingly innocuous genes. The identified variant, resulting in the substitution of histidine for aspartic acid at position 126, is located in a highly conserved region of the protein, suggesting its critical role in ribosome function.
The Functional Impact of the FEM1C Variant
The identified missense variant in FEM1C, substituting histidine for aspartic acid at position 126, is predicted to have a significant impact on protein function. This amino acid substitution occurs within a highly conserved region of the FEM1C protein, emphasizing its critical role in protein structure and function. To elucidate the functional consequences of this variant, in vitro studies were conducted to assess the protein's biochemical properties. The results demonstrated a marked reduction in the substrate binding affinity of the mutant FEM1C protein compared to the wild-type protein(Figure 1). This finding suggests that the variant protein is less efficient in interacting with its target molecules, which are essential for ribosome assembly. Given the pivotal role of the ribosome in protein synthesis, a compromised ability of FEM1C to participate in this process can have far-reaching implications for cellular function. A reduction in ribosome biogenesis or function can lead to a cascade of cellular defects, including decreased protein production, impaired cellular growth, and altered cellular signaling. These effects are particularly relevant in the context of neuronal development, where precise protein synthesis is essential for establishing neuronal connectivity, synaptic function, and overall brain development. Therefore, the observed reduction in FEM1C function provides a plausible molecular mechanism for the neurodevelopmental abnormalities observed in the patient.Furthermore, the variant protein may exhibit altered stability or protein-protein interactions, contributing to its impaired function. Additional studies are warranted to comprehensively characterize the functional consequences of this missense variant and to identify potential compensatory mechanisms.
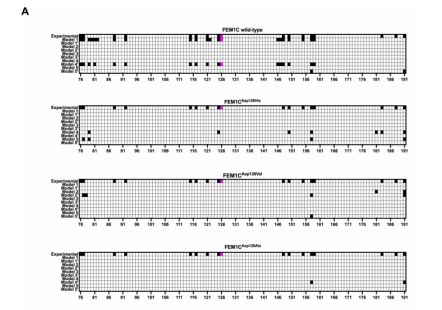
Figure 1.None of the mutants exhibited the peptide docked to its degron-binding pocket.
Neuronal Phenotype and Model Organism Studies
To investigate the potential impact of FEM1C dysfunction on neuronal development and function, the researchers utilized the nematode C. elegans as a model organism. This genetically tractable organism provides a robust platform for studying the complex interplay between genes and behavior. The choice of C. elegans is particularly relevant given the conserved nature of many core cellular processes, including protein synthesis and neuronal development, between nematodes and humans. Consistent with the patient's phenotype, fem-1 mutant worms exhibited impaired locomotion without overt muscle defects, strongly suggesting a primary defect in neuronal function(Figure 2). Detailed behavioral analysis revealed deficits in both spontaneous and evoked locomotion, indicating impaired motor coordination and responsiveness. To further elucidate the neuronal basis of the observed locomotor defects, the researchers performed detailed behavioral and physiological analyses. Mutant worms displayed increased sensitivity to the acetylcholinesterase inhibitor aldicarb, indicating a potential defect in cholinergic neurotransmission. Conversely, they exhibited normal responses to the acetylcholine receptor agonist levamisole, suggesting that the postsynaptic cholinergic system is largely intact(Figure 3).
These findings converge to support the hypothesis that FEM-1Asp133His-induced locomotor defects arise from synaptic dysfunction rather than muscle impairment. This aligns with the neurodevelopmental phenotype observed in the human patient, which includes pyramidal signs indicative of upper motor neuron dysfunction.
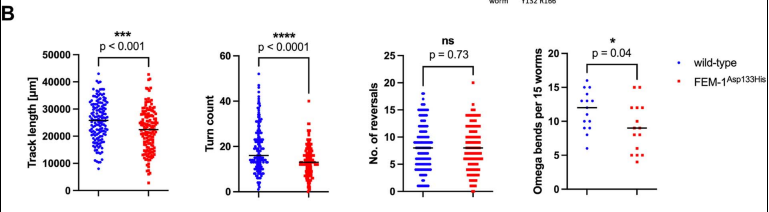
Figure 2. Track lengths, turn counts, a number of reversals, and omega bends in wild-type and FEM-1Asp133His mutants.
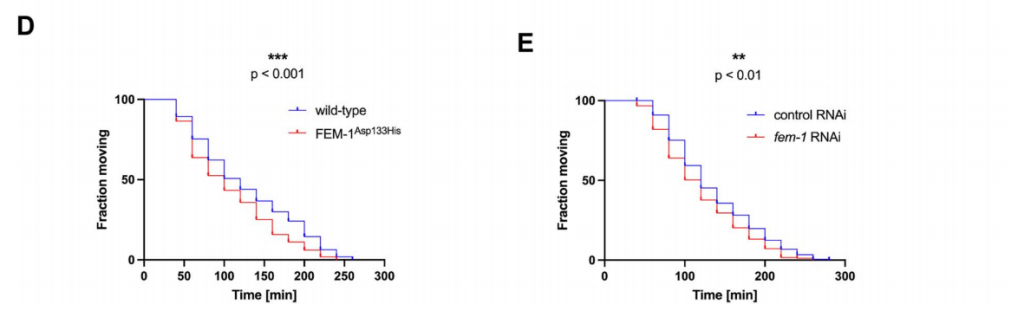
Figure 3. fem-1 mutants were significantly more sensitive to aldicarb but not to levamisole compared to wild-type worms. In addition, neuronal specific ribonucleic acid interference (RNAi) depletion of FEM-1 also sensitized animals to aldicarb treatment.
The implications of this study for the field of neurogenetics are profound. The identification of FEM1C as a causative gene for a severe neurodevelopmental disorder opens new avenues for research into the molecular mechanisms underlying this condition. Furthermore, it underscores the potential of utilizing model organisms, such as C. elegans, to rapidly translate findings from human genetics into functional insights. For our company, which specializes in C. elegans genome editing, this study presents an exciting opportunity. The identification of FEM1C as a key player in neurodevelopment provides a compelling target for further investigation. By leveraging our expertise in genome editing, we can create precise and efficient models of the human FEM1C variant in C. elegans to study the underlying molecular mechanisms in greater detail. Moreover, our platform can be utilized to screen for potential therapeutic compounds that may ameliorate the effects of FEM1C dysfunction. By identifying genetic suppressors or enhancers of the FEM1C phenotype, we can gain valuable insights into the molecular pathways involved and identify novel drug targets.
Reference:
Dubey AA, et al. A novel de novo FEM1C variant is linked to neurodevelopmental disorder with absent speech, pyramidal signs and limb ataxia.
Hum Mol Genet. 2023 Mar 20;32(7):1152-1161. doi: 10.1093/hmg/ddac276.
Keywords: FEM1C, neurodevelopmental disorder, absent speech, pyramidal signs, limb ataxia, protein synthesis, ribosome, genetic variant, C. elegans, genome editing





