Confirmation of the suitability of retron editing system for mammalian genome editing
2021-08-27 15:28
Bacteria have developed a subtle defense system to eliminate the foreign invasion genes of viruses and can quietly remove viral genes from their own genomes. This defense system of bacteria is a specific immunity system for archaea and bacteria to resist invasion by exogenous genetic material such as viruses. The currently popular CRISPR gene editing technology was developed from the bacterial defense system. Scientists are also delving into other defense systems of bacteria, such as the retron system, which has been parsed and applied in recent years.
The CRISPR system is highly targeted, but it is not very good at introducing new sequences into the target DNA. In contrast, retron can make a large number of copies of the target sequence by reverse transcriptase (RT), and these copies can be efficiently spliced into the host genome. By integrating retron with CRISPR, a new and more powerful gene editing tool can be developed. Recently, researcher Hui Yang and his colleagues demonstrated for the first time that this new gene editing tool, the retron system, can perform gene editing in mammalian cells.
Retron is composed of a specialized reverse transcriptase (RT) and a relevant non-coding RNA (ncRNA) that can be reversely transcribed to produce a multicopy single-stranded DNA (msDNA). The desired msDNA can be generated in vivo by replacing an insignificant region of the retron ncRNA with the desired sequence in gene editing (Fig. 1A). The investigators used four E. coli retrons from different branches (Ec48RT, Ec73RT, Ec86RT and Ec107RT) for gene editing and chose different combinations of coupling to verify the editing efficiency (Fig. 1B). These combinations include binding of retron RT to the amino terminus or carboxy terminus of Cas9 with XTEN linker (RT-Cas9 or Cas9-RT), and the fusion of retron ncRNA to the 5' or 3' end of gRNA (5' rgRNA or 3' rgRNA). In the relative abundance test of msDNA in human cells, retron RT combining with 3' rgRNA showed higher expression of msDNA comparing to that with 5' rgRNA (Fig. 1C). These results suggest that the retron RT has the same reverse transcription function in human cells.
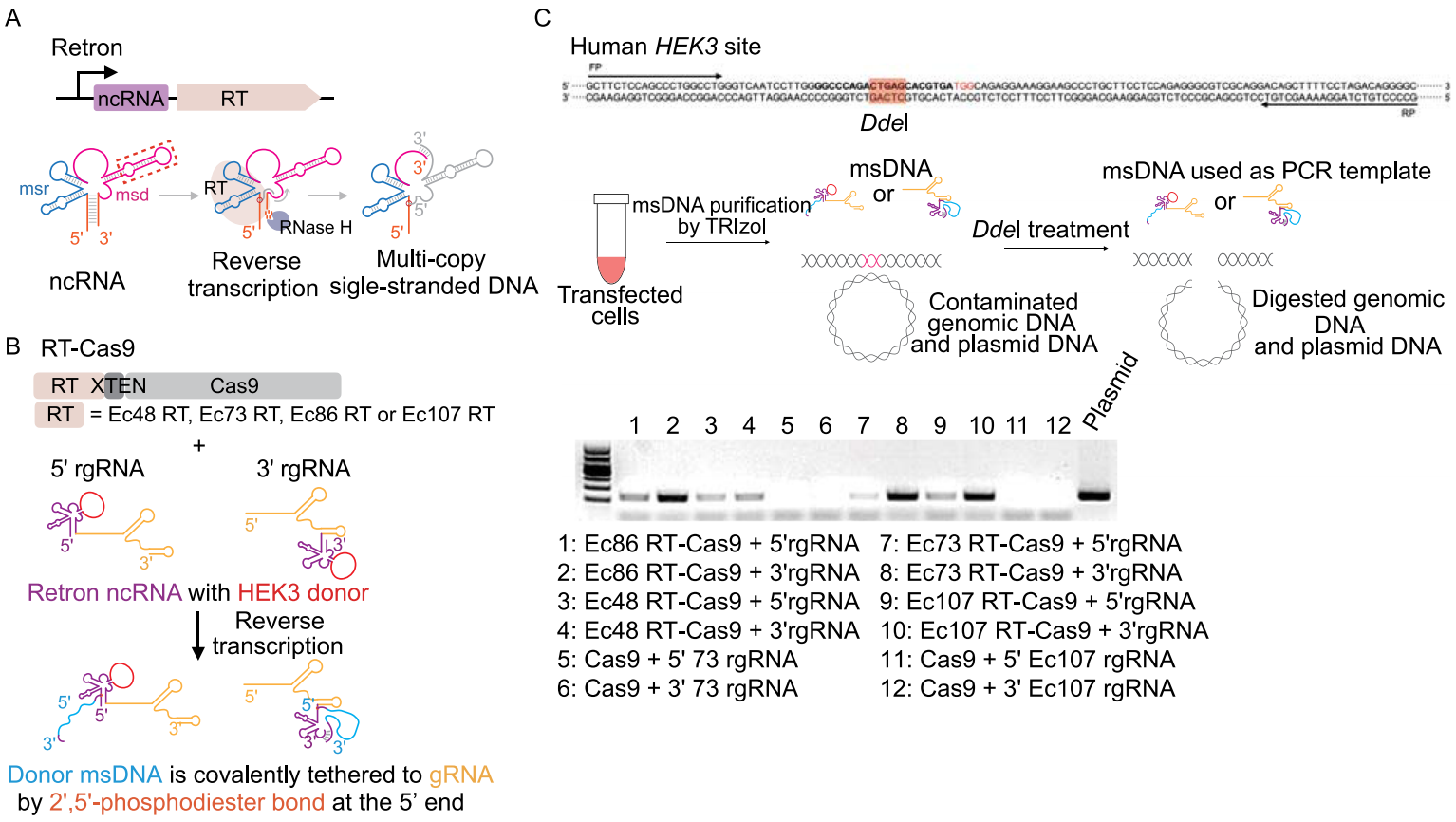
Figure 1. Retron-mediated generation of msDNA in human cells.
To confirm the applicability of the retron system in mammalian genome editing, the investigators transfected the human HEK293T cells with plasmids expressing different Cas9-RT fusions and verified the efficiency of retron editing-mediated HDR at EMX1 locus and HEK3 site by deep sequencing. Among four retrons tested, Ec86, Ec73 and Ec107 achieved varying degrees of precise editing and Ec73 combining with CRISPR-Cas9 showed highest activity, up to 10% (Fig. 2).
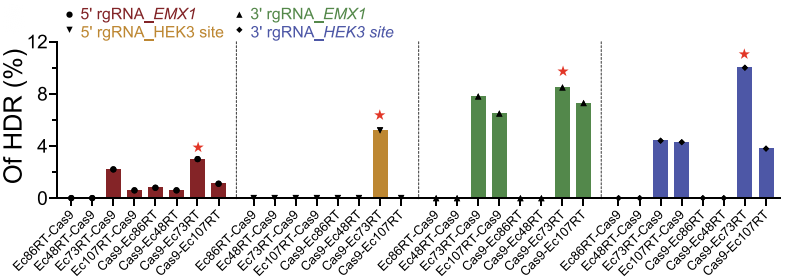
Figure 2. Analysis of retron editing efficiency by deep sequencing.
Compared with the more commonly used gene editing technologies, retron editing system relying on HDR mechanism is less restricted by PAM location and does not require additional exogenous donor template DNA. It is plausible to anticipate that retron editing system will be more accurate and safer due to the elimination of Cas9-induced DSBs, and is expected to be applied in the field of in vitro and in vivo gene editing therapy.
Reference: Kong, X., Wang, Z., Zhang, R. et al. Precise genome editing without exogenous donor DNA via retron editing system in human cells. Protein Cell (2021).
DOI: https://doi.org/10.1007/s13238-021-00862-7




