C. elegans Cell Invasion Model Helps Study of Cancer Cell Metastasis Mechanism
2020-03-25 17:20
Cancer has become one of the serious threats to human health, and its treatment has become the focus of global medical strategy. Cancer metastasis is an important reason for its high lethality. Generally, cancer cells will normally proliferate or invade bodies[1], and enhanced cell invasiveness is a feature of cancer metastasis[2]. Cancer cells metastasis begins from invading and penetrating through the basement membrane to get into body cavities, blood, etc., and then invading new tissues and organs, such as the liver, brain, and lungs, destroying its functions and eventually causing death. Therefore, understanding the mechanism of cancer cells metastasis can offer important targets for the development of new drugs to inhibit cancer metastasis.
The first step of invasion is that cancer cells pass through the basement membrane, which often occurs in the deep tissue layer and is difficult to be visualized. SunyBiotech’s client, professor David Q. Matusand and his team, of State University Of New York-Stony Brook, intelligently utilizing the morphogenesis of C. elegans uterus-- Vulvar connection morphogenesis as an easy to operate and easy to observe model to study cell invasion in the body[3]. The cells that can invade the basement membrane in this model are called anchor cells. The team found that during the development of nematodes, there exists a complex gene regulatory network to ensure the normal differentiation and invasion of anchor cells.
Anchor cells are generated in the L2 stage, when two initially equal cells divide through a random asymmetric gap, generating terminally differentiated anchor cells AC and proliferating ventral (VU) cells[4]. In the middle of L3 stage, anchor cells invade the basal layer and connect the uterus and vulvar epithelium to promote spawning[3]. Anchorage differentiation is formed by the regulation of multiple transcription factors mediated by the Notch signaling pathway. Previous studies identified four transcription factors, fos-1a, egl-43, hlh-2, and nhr-67 to mediate anchor cells differentiation and invasion, but how to regulate both differentiation and invasion is still unclear (Figure 1).
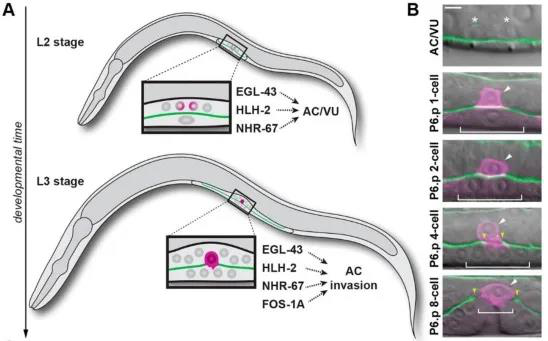
Figure 1: Transcription factors regulate AC cell differentiation and invasion
The authors used RNAi interference technology to interfere with the expression of these transcription factors and observed their effects on AC cell differentiation and invasion. In addition, the authors also adapted CRISPR / Cas9 gene editing technology to label these transcription factors with fluorescent proteins, including the green fluorescent protein labeled nhr-67 (syb509) which ordered from SunyBiotech, so that the interaction between these transcription factors during differentiation and invasion can be studied by observing the expression of fluorescent protein. It was found that before differentiation of AC / VU , egl-43, hlh-2 and nhr-67 independently regulated the expression of endogenous lin-12 (Notch's homologous gene in nematodes) genes and promoted differentiation of anchor cells. After anchor cell differentiation, egl-43, hlh-2, and nhr-67 function in parallel with fos-1 in a positive feedback loop, promoting AC cells to invade the basement membrane (Figure 2).
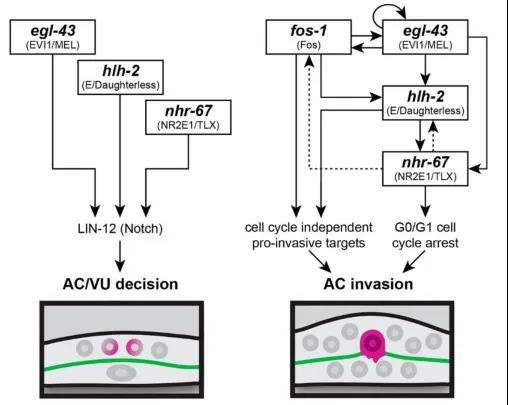
Figure 2: Transcription factors regulation co-regulates AC differentiation and invasion
In summary, transcription factors independently regulate the differentiation of AC / VU anchor cells mediated by Notch signals, and then a gene regulatory network can be quickly assembled to enhance AC invasion.
If you have a need for customized C. elegans gene editing, please feel free to contact SunyBiotech!
Reference :
1. Kohrman, A. Q. and Matus, D. Q. (2017). Divide or Conquer: Cell Cycle Regulation of Invasive Behavior. Trends Cell Biol. 27, 12–25.
2. Hanahan, D. and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144, 646–674.
3. Sherwood, D. R. and Sternberg, P. W. (2003). Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell 5, 21–31.
4. Wilkinson, H. A., Fitzgerald, K. and Greenwald, I. (1994). Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79, 1187–1198.
Link:https://www.biorxiv.org/content/10.1101/691337v3




